Random Forest Regressor
Random forest regressor is a variant of the random forest classifier.
It is primarily used for classification tasks. This model is an ensemble of decision trees.
It combines the predictions of multiple individual trees to imrpove performance.
By aggregating the results from those trees, typically through votng or avaeraging.
It produces a final prediction that is more accurate than any single tree alone.
we import sns and pd from the necessary libraries.
import seaborn as sns
import pandas as pd
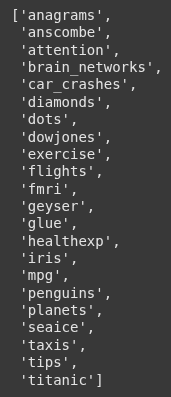
Next we call sns.get_dataset_names() to get the list of all the buit-in datasets available in the Seaborn library.
sns.get_dataset_names()
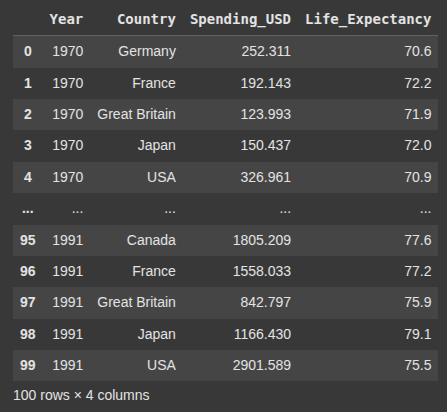
Next we load the ‘healthexp’
healthexp = sns.load_dataset('healthexp')
healthexp.head(100)
here we convert categorical columns in the heathexp DataFrame into numerical format using what is known as one-hot encoding.
healthexp = pd.get_dummies(healthexp)
Next we remove the Life_Expectancy column from the healthexp DataFrame and storing the remaining coluns in X
X = healthexp.drop(['Life_Expectancy'], axis=1)
we then assign Life_Epectancy colunm from the healthexp DataFrame to the variable y, which represents the target variable in a machine learning task.
y = healthexp['Life_Expectancy']
Next we import train_test_split and pass our X and y to it, including the test size 0.2(20% of the data), and a random_state =19 for reproducibility
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2, random_state=19)
Then we import the model RandomForestRegressor and assign the variable “rfr” to it.
from sklearn.ensemble import RandomForestRegressor
rfr = RandomForestRegressor(random_state=13)
Next we train the model using the .fit()
rfr.fit(X_train, y_train)
Then we evaluate the model using the .predict() and passing the X_test.
We do this to measure what the model have learnt.
y_pred = rfr.predict(X_test)
Next we import our evaluation metrics.
mean_absolute_error:Measures the average absolute difference between actual and predicted values.
mean_squared_error:Measures the average of the squared differences.
r2_score: Measures how well the predictions approximate the actual values..
from sklearn.metrics import mean_absolute_error, mean_squared_error, r2_score
mean_absolute_error(y_test, y_pred)
mean_squared_error(y_test, y_pred)
r2_score(y_test, y_pred)
Read more on it..
Here we perform hyperparameter tuning.
This means we are tuning the hyperparameters to enable us have better prediction or to enable the modle learn better.
param_grid = {
'n_estimators': [100, 200, 300], # Number of trees in the forest
'max_depth': [ 10, 20, 30], # Maximum depth of the tree
'min_samples_split': [2, 5, 10], # Minimum number of samples required to split an internal node
'min_samples_leaf': [1, 2, 4], # Minimum number of samples required to be at a leaf node
}
Here we imports GridSearchCV, a utility from scikit-learn used to automatically search for the best hyperparameters for a given model.
from sklearn.model_selection import GridSearchCV
The we pass our model “rfr, and he hyperparameters to the GridSearchCV
rfr_cv = GridSearchCV(estimator=rfr, param_grid=param_grid, cv=3, scoring='neg_mean_squared_error', n_jobs=-1)
Then we train using the .fit()
rfr_cv.fit(X_train, y_train)
Next we evaluate or test the performance of our model.
amd also perform evaluation metrics to see if it performs better than the previous training.
y_pred = rfr_cv.predict(X_test)
mean_absolute_error(y_test, y_pred)
mean_squared_error(y_test, y_pred)
r2_score(y_test, y_pred)
Ryan is a Data Scientist at a fintech company, where he focuses on fraud prevention in underwriting and risk. Before that, he worked as a Data Analyst at a tax software company. He holds a degree in Electrical Engineering from UCF.
